The general understanding of fertilization is the fusion of two haploid gametes, the spermatozoon, and the oocyte, to form a diploid cell that goes through the multiple cell division and cell differentiation events of embryo development. However, recent research has pushed this understanding.
Parthenogenesis, also known as virgin births, is another way of embryo development without fertilization of the oocyte by a spermatozoon. Hence, there is no genetic contribution from a male gamete. Parthenogenesis is observed in many invertebrates—rotifers, snails, water fleas, aphids, zooplankton, and some honeybees. A few cases of parthenogenesis in some sharks, salamanders, lizards, and snakes have also been observed. During natural parthenogenesis, the oocyte either duplicates its genetic material without a cell division or by incorporating the polar body that contains the other half of the genetic material expelled in the meiotic process. Consequently, parthenogenesis will only result in female offspring. Parthenogenesis has been mimicked artificially. However, these studies have highlighted the essential aspect of the fine coordination of the male and female genome during mammalian development, as most embryos die during the gestation period. In mammals, this is coordinated by the maternal-specific imprinting of the diploid genome. Imprinting is a process that orchestrates the correct combination of the maternal and paternal contribution in the zygote by adding or removing methyl groups to specific genomic regions.
Recently, a publication in PNAS described a process of creating viable offspring from single unfertilized mouse oocytes [1]. Wei and colleagues tested the importance of different imprinting regions found in the literature for the generation of offspring. Using the B6CASTF1 mouse strain (C57BL/6 ♀ × CAST ♂), in which the paternal and maternal copies of the genome easily can be distinguished, they created parthenogenic embryos by changing the methylation pattern in a targeted allele-specific manner with the help of single-guide RNAs (sgRNAs). The applied sgRNAs are RNA molecules with dual functions; one part targets the specific DNA region, while the other part is responsible for the methylation editing. Wei and colleagues identified several regions where targeted methylation editing affected embryonic, fetal, and/or postnatal growth and development. Altogether, they were able to determine a combination of 7 regions necessary for the generation of viable full-term offspring from single unfertilized mouse oocytes.
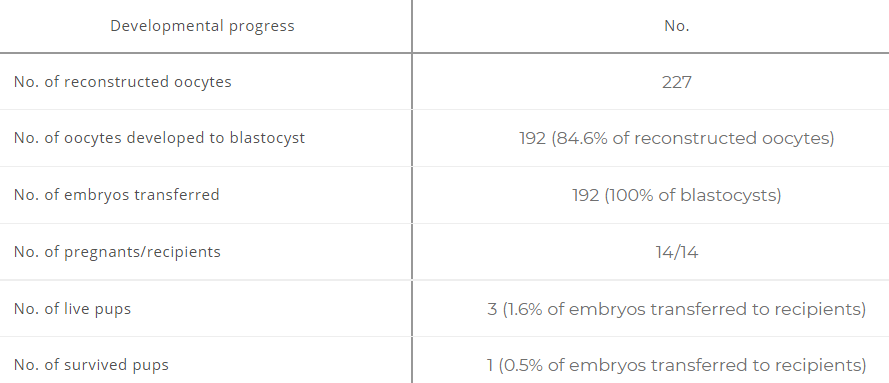
This study demonstrates that parthenogenesis can be achieved in mammals, but where does this study leave the male contribution in the generation of offspring?
Men, you are not dispensable yet. There are several other aspects to be considered before this technique can be possible in humans, e.g., there are clear roles of the sperm-derived RNAs on embryo development; human oocytes, unlike mice, cannot undergo early embryonic divisions in the absence of sperm-derived centrioles, etc. It will be interesting to follow how this knowledge can be applied in the coming years in agriculture, research, and medicine.
[1] Wei Y, Yang C, Zhao Z (2022). Viable offspring derived from single unfertilized mammalian oocytes. PNAS, 119. DOI: 10.1073/pnas.2115248119
