Preface :
Dear Member,
In order to dynamize the website we will in the future upload the different parts of our newsletter as they are finished. In the end of the season we will post the full newsletter as usual.
Please find our first part of the autumn newsletter – an interview with Dr. Frederic Chalmel.
Enjoy your reading!
 Tête-à-tête with Frédéric Chalmel
Tête-à-tête with Frédéric Chalmel
 Dr. Frédéric Chalmel is a research associate at IRSET institut and holds a PhD in Bioinformatics. With 15 years of experience in functional genomics, his current research focuses on the “omic-biology” of mammalian spermatogenesis with emphasis on transcriptomic and integrative genomics. He deploys integrative genomics methodologies applied to reproductive biology and reproductive toxicology. Specifically, he investigates the effects of xenobiotics on transcriptional and epigenetic dynamics during male germ cell differentiation as well as gonad development in human. In 2015 Frederic/Dr. Chalmel was awarded our young research prize for his work on the complex and highly coordinated molecular networks underlying normal and pathological spermatogenesis.
Dr. Frédéric Chalmel is a research associate at IRSET institut and holds a PhD in Bioinformatics. With 15 years of experience in functional genomics, his current research focuses on the “omic-biology” of mammalian spermatogenesis with emphasis on transcriptomic and integrative genomics. He deploys integrative genomics methodologies applied to reproductive biology and reproductive toxicology. Specifically, he investigates the effects of xenobiotics on transcriptional and epigenetic dynamics during male germ cell differentiation as well as gonad development in human. In 2015 Frederic/Dr. Chalmel was awarded our young research prize for his work on the complex and highly coordinated molecular networks underlying normal and pathological spermatogenesis.
When and why did you decide to work in Male Fertility?
It was a combination of circumstances. In 2005, after my PhD thesis, I was looking for a post-doc working in the genomic field. I found a position at the Biozentrum (Basel, Switzerland) in the transcriptomic platform headed by Dr. Mike Primig, an expert in the genomics of yeast meiosis. The aim of my researches was to decipher the gene expression program of male germ cells in mammals in close collaboration with Pr. Bernard Jégou (IRSET, Rennes, France). Quickly, I realized how interesting spermatogenesis was. There were and there still are so many things to explore and to discover about the germline directly linked to critical questions related to evolution, species perpetuation and thus to male fertility.
In 2015, you received the INYRMF award, what did this award bring you?
I would be lying if I said that I wasn’t proud of this award. Being awarded in 2015 ranks right up there – to be recognized by my peers is validation of the work and contributions I’ve made in this field over the last decade. This demonstrates that (reproductive) genomics, just as molecular biology few decade decades ago, is now widely accepted as another complementary tool added to our arsenal for studying reproductive biology.
Can you name the greatest success(es) in your career?
Careers in research in the French/European Research Area have become so difficult that every grant we get, every articles we publish are success. Instead of talking about success(es) I would prefer to mention the greatest joy(s) of my career and there are so numerous. First, at the human level, I’m so lucky to work with my teammates in a friendly and dynamic ambiance but at the same time serious and of a high intellectual level. Dr Antoine Rolland and I try everything to maintain this atmosphere in our research group. Another great happiness is also all opportunities I got to meet and collaborate with other great researchers from other labs. At the scientific level, Pr. Bernard Jégou and I recently co-supervised a study that could be considered as one of the first in the field of paleo-infertility and it was one of the most stimulating experience in my career.
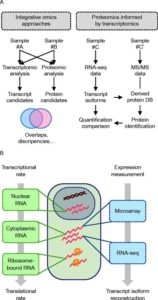 When transcriptomics meets proteomics. (A) Typical integrative omics approaches involve the combination of datasets originating from various technologies, most notably transcriptomics and proteomics. Such strategies are often used to identify more reliable candidates (i.e. factors evidenced at both the RNA and protein levels), but they can also be useful in order to compare and correlate transcription and translation rates. More recently, the combination of RNA-seq and mass spectrometry (MS)-based proteomic has led to the development of the so-called proteomics informed by transcriptomics (PIT) approach. In this approach, the protein sequence database (DB) queried for protein identification purpose is directly derived from transcript sequences obtained following RNA-seq analysis of the same or equivalent sample as that used for MS/MS analysis. (B) The characterization of the transcriptome has long been used as a proxy for the proteome. However, depending on whether nuclear, total cytoplasmic, or ribosome-bound RNAs are analysed, the captured picture will reflect either more the transcriptional rate or the translational rate. Additionally, while both approaches perform equivalently from a quantitative point of view, RNA-seq overcomes microarray technology from a qualitative point of view as it allows full-length transcript reconstruction and can thus discriminate between distinct protein-coding isoforms.
When transcriptomics meets proteomics. (A) Typical integrative omics approaches involve the combination of datasets originating from various technologies, most notably transcriptomics and proteomics. Such strategies are often used to identify more reliable candidates (i.e. factors evidenced at both the RNA and protein levels), but they can also be useful in order to compare and correlate transcription and translation rates. More recently, the combination of RNA-seq and mass spectrometry (MS)-based proteomic has led to the development of the so-called proteomics informed by transcriptomics (PIT) approach. In this approach, the protein sequence database (DB) queried for protein identification purpose is directly derived from transcript sequences obtained following RNA-seq analysis of the same or equivalent sample as that used for MS/MS analysis. (B) The characterization of the transcriptome has long been used as a proxy for the proteome. However, depending on whether nuclear, total cytoplasmic, or ribosome-bound RNAs are analysed, the captured picture will reflect either more the transcriptional rate or the translational rate. Additionally, while both approaches perform equivalently from a quantitative point of view, RNA-seq overcomes microarray technology from a qualitative point of view as it allows full-length transcript reconstruction and can thus discriminate between distinct protein-coding isoforms.
From: Chalmel F, and Rolland AD (2015). Linking transcriptomics and proteomics in spermatogenesis. Reproduction, 150: 149-57. doi: 10.1530/REP-15-0073.
Can you name a moment of failure (and which lesson did you learn with it)?
In science, success(es) and failure(s) are indissociable and the latter is an important and inevitable aspect of our work. There are also other kind of failures. I would say that the worst thing which frequently happens nowadays in our lab is that we are forced to separate us from young and highly competent scientists just because of administrative reasons. Unfortunately, I have never learned from this experience and it’s always a though time.
Which advice(s) would you give to young researchers in Male Fertility?
Always keep an open mind, never stop to learn and avoid any a priori, there are absolute prerequisites in our profession if you want to keep the spark of creativity alive and to open up new opportunities.


